massXpert2 User Manual
8 Data Customizations
In this chapter, the user will be walked trough an example of how new polymer chemistry definition data can be generated and included in the automatic “data detection system” of massXpert (that is how new polymer chemistry definitions should be registered with the system).
Customization is typically performed by the normal user (not the
Administrator nor the Root of the machine) and as such new data are
typically stored in the user's “home” directory. On
UNIX machines, the “home” directory
is usually the /home/username
directory, where username is the login username. On
MS-Windows, that directory is typically the
C:/Documents and Settings/username
once again with username being the login username.
Tip
Although MS-Windows pathnames use a back slash (“\”, in this book these are composed using forward slashes for a number of valid reasons. The reader only needs to replace back slashes with the forward variety.
In the next sections we will refer to that “home directory” (be it on UNIX or MS-Windows machines) as the $HOME directory, as this the standard environment variable describing that directory in GNU/Linux.
When massXpert is executed, it automatically tries to read data
configuration files from the home directory (in the .massXpert2 directory). Once this is done, it
reads all the data configuration files in the installation directory
(typically, on GNU/Linux that would be the
configuration data in the /usr/share/massxpert2/data directory or, on
MS-Windows, the c:/Program Files/massXpert2/data directory).
We said above that massXpert2 tries to read the data configuration files from the home directory. But upon its very first execution, right after installation, that directory does not exist, and in fact massXpert2 creates that directory for us to populate it some day with interesting new data.
The $HOME/massxpert2 directory
should have a structure mimicking the one that was created upon installation
of the software, that is, it should contain the following two directories:
polChemDefspolSeqs
Those are the directories where the user is invited to store their
personal data. In order to start a new definition, one might simply copy in
the polChemDefs one of the polymer
chemistry definitions that are shipped with massXpert. What should be copied?
An entire polymer chemistry definition directory, like for example the
following:
/usr/local/share/massxpert2/polChemDefs/protein-1-letter
or
C:/Program
Files/msXpertSuite/data/massxpert2/polChemDefs/protein-1-letter
Once that polymer chemistry definition is copied, one may start studying how it actually works. This directory contains the following kinds of files:
protein-1-letter.xml: the polymer chemistry definition file. This is the file that is read upon selection of the corresponding polymer chemistry definition name in XpertDef. If the polymer chemistry definition is not yet registered with the system (described later), then open that file by browsing to it by clicking Cancel (see Chapter 3, XpertDef: Definition of Polymer Chemistries);SVG files: scalar vector graphics files used to render graphically the sequence in the sequence editor. For example,
arginine.svgcontains the graphical representation of the arginine monomer. There are such graphics files also for the modifications (like, for example, thesulphation.svgcontains the graphical representation of the sulphation modification. Figure 8.1, “The polymer chemistry definition directory” shows two examples of SVG files belonging to two distinct polymer chemistry definitions;chemPad.conf: configuration file for the chemical pad in the XpertCalc module;monomer_dictionary: file establishing the relationship between any monomer code of the polymer chemistry definition and the graphical SVG file to be used to render graphically that monomer in the sequence editor;modification_dictionary: file establishing the relationship between any monomer modification (see Section 4.8.1, “Selected Monomer(s) Modification”) and the graphical SVG file to be used to render graphically that modification onto the modified monomer in the sequence editor;cross_linker_dictionary: file establishing the relationship between any cross-link (see Section 4.9, “Monomer Cross-linking”) and the graphical SVG file to be used to render graphically that cross-link onto the cross-linked monomers in the sequence editor;pka_ph_pi.xml: file describing the acido-basic data (see Section 4.16, “pKa, pH, pI and Charges”) pertaining to ionizable chemical groups in the different entities of the polymer chemistry definition;
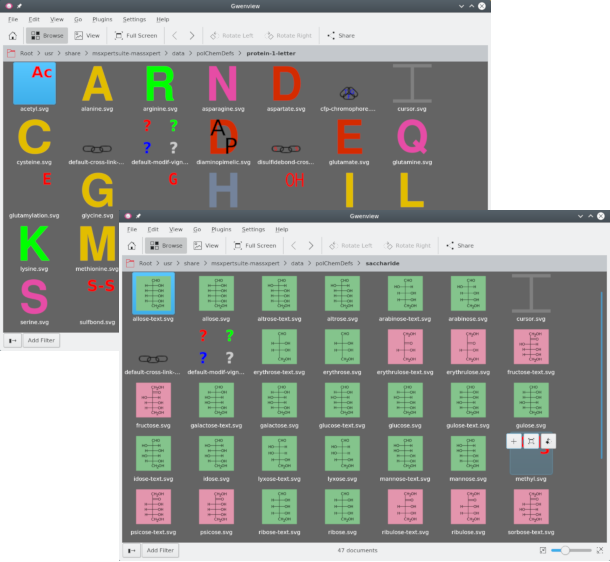
Each monomer of the polymer chemistry definition ought to have a corresponding SVG file with which it has to be rendered graphically should that monomer be inserted in the polymer sequence. This example shows two SVG files corresponding to two monomers each belonging to a different polymer chemistry definition.
Figure 8.1: The polymer chemistry definition directory #
The polymer sequence editor is not a classical editor. There is no font in this editor: when the user starts keying-in a polymer sequence in the editor, the small SVG graphics files are rendered into raster vignettes at both the proper resolution and screen size and displayed in the sequence editor. The user is totally in charge of designing the SVG graphics files for each of the monomers defined in the polymer sequence editor. Of course, reusing material is perfectly possible.
There is one constraint: that the
monomer_dictionary file lists with precision
“what monomer code goes with what SVG graphics
file”. That file has the following contents, for example, for the
“protein-1-letter” polymer chemistry definition, as shipped
in the massXpert package:
# This file is part of the massXpert2 project.
# The "massXpert2" project is released ---in its entirety--- under the
# GNU General Public License and was started (in the form of the GNU
# polyxmass project) at the Centre National de la Recherche
# Scientifique (FRANCE), that granted me the formal authorization to
# publish it under this Free Software License.
# Copyright (C) 2006,2023 Filippo Rusconi
# This is the monomer_dictionary file where the correspondences
# between the codes of each monomer and their graphic file (pixmap
# file called "image") used to graphicallly render them in the
# sequence editor are made.
# The format of the file is like this :
# -------------------------------------
# A%alanine.svg
# where A is the monomer code and alanine.svg is a
# resolution-independent svg file.
# Each line starting with a '#' character is a comment and is ignored
# during parsing of this file.
# This file is case-sensitive.
A%alanine.svg
C%cysteine.svg
D%aspartate.svg
E%glutamate.svg
F%phenylalanine.svg
G%glycine.svg
H%histidine.svg
I%isoleucine.svg
K%lysine.svg
L%leucine.svg
M%methionine.svg
N%asparagine.svg
P%proline.svg
Q%glutamine.svg
R%arginine.svg
S%serine.svg
T%threonine.svg
V%valine.svg
W%tryptophan.svg
Y%tyrosine.svg
What one sees from the contents of the file is that each monomer code
has an associated SVG file. For example, when the user
has to key-in a valine monomer, they key-in the code V and
XpertEdit knows that the monomer vignette to show has to be rendered using the
valine.svg file.
For the monomer modification graphical rendering, the situation is
somewhat different, as seen in the
modification_dictionary file:
# This file is part of the massXpert2 project.
# The "massXpert2" project is released ---in its entirety--- under the
# GNU General Public License and was started (in the form of the GNU
# polyxmass project) at the Centre National de la Recherche
# Scientifique (FRANCE), that granted me the formal authorization to
# publish it under this Free Software License.
# Copyright (C) 2006,2023 Filippo Rusconi
# This is the modification_dictionary file where the correspondences
# between the name of each modification and their graphic file (pixmap
# file called "image") used to graphicallly render them in the
# sequence editor are made. Also, the graphical operation that is to
# be performed upon chemical modification of a monomer is listed ('T'
# for transparent and 'O' for opaque). See the manual for details.
# The format of the file is like this :
# -------------------------------------
# Phosphorylation%T%phospho.svg
# where Phosphorylation is the name of the modification. T indicates
# that the visual rendering of the modification is a transparent
# process (O indicates that the visual rendering of the modification
# is a full image replacement 'O' like opaque). phospho.svg is a
# resolution-independent svg file.
# Each line starting with a '#' character is a comment and is ignored
# during parsing of this file.
# This file is case-sensitive.
Phosphorylation%T%phospho.svg
Sulphation%T%sulpho.svg
AmidationAsp%O%asparagine.svg
Acetylation%T%acetyl.svg
AmidationGlu%O%glutamine.svg
Oxidation%T%oxidation.svg
There are two ways to render a chemical modification of a monomer:
Opaque rendering: the initial monomer vignette is replaced using the one listed in the file for the modification. One example is the “AmidationGlu” modification:
AmidationGlu%O%glutamine.svg
Upon amidation of a glutamyl residue (“AmidationGlu” is the name of a modification in the current polymer chemistry definition), the graphical representation of the modification involves the replacement of the glutamyl residue vignette in the sequence editor with the new one, that happens to be in the
glutamine.svgfile. In other words, the process involves an “Opaque” overlay of the vignette for unmodified Glu with a vignette rendered by using theglutamine.svgfile.Transparent rendering: the initial monomer vignette is overlaid with a new vignette that is read from an SVG file that has a transparent background. One example is the “Phosphorylation” modification:
Phosphorylation%T%phospho.svg
The monomer undergoing a phosphorylation has its vignette overlaid with a “Transparent” one showing a small red “P” that is read from the
phospho.svgfile.
When designing vignettes, the best thing to do is to convert the text to path, so that the rendering is absolutely perfect.
Warning
It is absolutely essential, for the proper working of the sequence editor, that the SVG files be square (that is, width = height).
Once the new polymer chemistry has been correctly defined, it is time to register that new definition to the system. To recap: all the files for that definition should reside in a same directory, exactly the same way as the files pertaining to a given polymer chemistry definition are shipped in massXpert altogether in one directory. The name of the new polymer chemistry definition should be unambiguous, with respect to other registered polymer chemistry definitions.
The way personal polymer chemistry definitions are registered is by creating a personal polymer chemistry definition catalogue file, which must comply with both following requirements:
Be named
xxxxx-polChemDefsCat, with “xxxxx” being a discretionary string (this might well be your login username). The requirement is that-polChemDefsCatbe the last part of the filename.
Tip
Please DO NOT USE spaces or diacritical signs in your filenames. RESTRICT yourself to ASCII characters between [a-z], [0-9], “_” and “-”.
This is actually something very general as a recommendation in order to not suffer from severe headaches when you least expect them…
Be located in the
$HOME/.massXpert2/data/polChemDefsdirectory and have the following format:dna=/path/to/definition/directory/dna/dna.xml
In this example, the “dna” polymer chemistry definition is being registered as a file
dna.xmllocated in thednadirectory, itself located in the/path/to/definition/directorydirectory;
Note that if a new polymer chemistry definition should be made available system-wide, then it is logical that its directory be placed along the ones shipped with massXpert and a new local catalogue file might be created to register the new polymer chemistry definition.
At this point the new polymer chemistry definition might be tested. Typically, that involves restarting the massXpert program and creating a brand new polymer sequence of the new definition type. The first step is to check if the new definition is successfully registered with the system, that is, it should show up a an available definition upon creation of the new polymer sequence. If not, then that means that the catalogue file could not be found or parsed correctly.
When problems like this one occurs, the first thing to do is to ensure that the console window (on MS-Windows it is systematically started along with the program; on GNU/Linux the way to have it is to start the program from the shell) so as to look with attention at the different messages that might help understanding what is failing.
Please, do not hesitate to submit bug reports (see Section 2, “Feedback from the users” for the method to contect the author for feature requests or bug reports).
