MassXpert3 User Manual
- Preface
- 1 Generalities
- 2 Basics in Polymer Chemistry
- 3 XpertDef: Definition of Polymer Chemistries
- 4 XpertEdit: a Powerful Editor and Simulation Center
- 5 XpertCalc: a Powerful Mass Calculator
- 6 XpertMiner: a Data Miner
- 7 XpertScript: a Scripting Console
- 8 Utilities
- 9 Data Customizations
- A GNU General Public License version 3
7 XpertScript: a Scripting Console
XpertScript is a module that has been conceived as a scripting console allowing the user to automate a number of tasks.
7.1 XpertScript Invocation #
The XpertScript module is easily called by pulling down the menu item from the MassXpert program's menu. Clicking on → will open the scripting console window, as represented in Figure 7.1, “The scripting console window” .
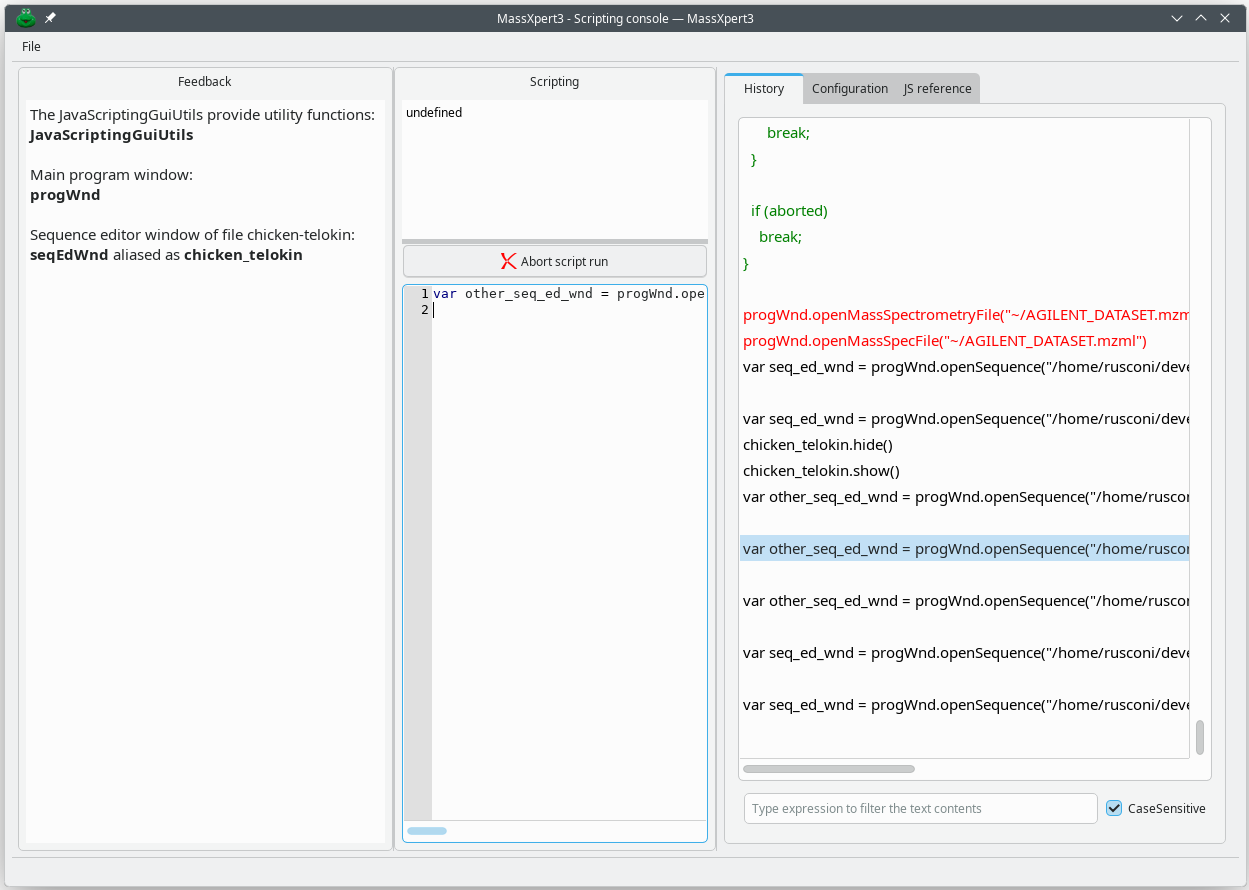
Scripting console window.
The window is divided into three vertical panes. The left hand side pane collects all the program's feedback to the user, so they are informed of any new variable that is created in the JavaScript environment and made available to them. The middle pane has two regions. The upper region will display all the return values after any JavaScript command has been run. The lower region is the actual script input region. The right hand side region contains mainly the history of the commands issued during the previous and current scripting sessions.
Figure 7.1: The scripting console window #
7.2 Running Scripts in the Scripting Console #
The lower part of the middle panel of the scripting window is a text editor that has syntax colouring for JavaScript code. After having entered the script, hitting Enter while maintaining Ctrl pressed will run the script.
Upon running of the script, the output of the script is printed in the upper part of the same middle panel. Most often the output will be undefined (in grey colour), which is just fine. If the output is some actual value, it will be printed in green. If there was an error, the ouptut will be printed in red.
Note that each time a script is run, it is appended in the right hand side panel of the window, in the the History tab. The coloring of the items added to the History> tab follows the same logic as that described above.
7.3 Loading Script Files in the Scripting Console #
It is possible to craft ECMAscript-compliant scripts outside of the JavaScript console window, store them in files and open them for running in the JavaScript console window. To do so, select → .
Once the script has been inserted into the script editor widget, the user can run it by hitting Enter while maintaining Ctrl pressed.
7.4 One Elaborate Example of Script #
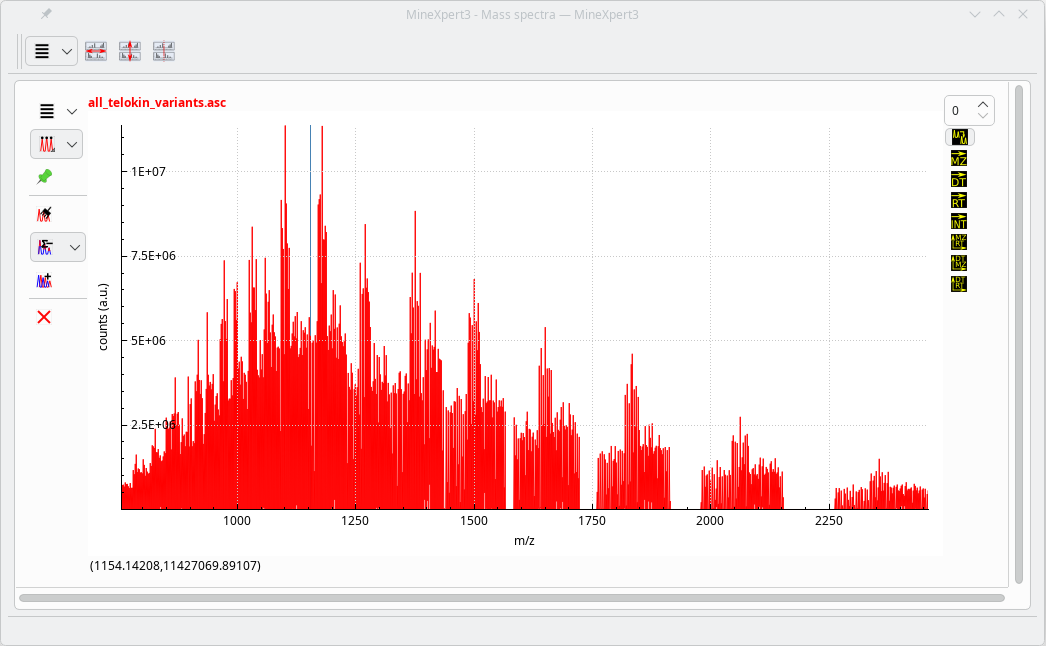
The following script was used to generate a mass spectrum that represents the full molecular diversity of a protein from chicken: Telokin , the sequence of which is shipped along MassXpert in the sample sequences.
Telokin has been the subject of the author's Ph.D. research work and the deciphering of its incredible molecular diversity was published in Biochemistry in 1997. The author gives classes of mass spectrometry for biology, and that protein is the subject of the hands-on session of these classes.
function getRandomWithinPercent(value, percent)
{
// Calculate the % range
const range = value * percent;
// Calculate min and max bounds
const min = value - range;
const max = value + range;
// Generate random number within the range
return Math.random() * (max - min) + min;
}
var seq_ed_wnd = progWnd.openSequence("chicken-telokin.mxp");
polymer = seq_ed_wnd.getPolymer();
polymer.setLeftEndModifByName("Acetylation");
let phosphoserine_index = 12;
polymer.modifyMonomer(phosphoserine_index, "Phosphorylation",
/*override*/ false);
calc_options = polymer.getCalcOptions();
ionizer = polymer.getIonizer();
seq_ed_wnd.setTraceColor("red");
mass_peak_shaper_dlg = seq_ed_wnd.configureMassPeakShaper();
mpscw = mass_peak_shaper_dlg.getConfigWidget();
let Mr = 17336;
let z_dict = {
7: 0.125e6,
8: 0.25e6,
9: 0.375e6,
10: 0.5e6,
11: 0.675e6,
12: 0.876e6,
13: 0.9e6,
14: 1e6,
15: 1e6,
16: 0.875e6,
17: 0.75e6,
18: 0.5e6,
19: 0.375e6,
20: 0.25e6,
21: 0.125e6
};
// let z_dict = { 1: 1e7 }
let random_intensity = 0
let nterm_indices_list = [1, 2, 7];
// let nterm_indices_list = [0]
aborted = false;
for (const nterm_index of nterm_indices_list)
{
// let removed_glu_start = 0
// let removed_glu_stop = 1
let removed_glu_start = 1;
let removed_glu_stop = 7;
for (let removed_glu = removed_glu_start; removed_glu <
removed_glu_stop; removed_glu++)
{
let start_index = nterm_index;
let end_index = polymer.size - 1 - removed_glu;
range_as_string = `[${start_index}-${end_index}]`;
seq_ed_wnd.setIndexRanges(range_as_string,
libXpertMassCore.Enums.LocationType.INDEX);
let mz = 1;
for (let z in z_dict)
{
if(checkAbort())
{
aborted = true;
break;
}
intensity = z_dict[z];
if (z == 0)
{
mz = Mr;
}
else
{
mz = Mr / z;
}
ionizer.level = z;
polymer.setIonizer(ionizer);
mass_peak_shaper_dlg.setParameters(mz, 150,
libXpertMassCore.Enums.MassPeakShapeType.GAUSSIAN, 40000, 0, 6);
mpscw.checkParameters();
// Now perform two calculations: without or with the polymer acetylation
calc_options.setPolymerEntities(libXpertMassCore.Enums.ChemicalEntity.NONE);
seq_ed_wnd.setCalcOptions(calc_options)
random_intensity = getRandomWithinPercent(intensity, 0.10);
seq_ed_wnd.setNormalizationIntensity(random_intensity);
seq_ed_wnd.synthesizeMassSpectra();
// Now perform the calculation with the Phosphorylation.
calc_options.setMonomerEntities(libXpertMassCore.Enums.ChemicalEntity.MODIF);
seq_ed_wnd.setCalcOptions(calc_options);
random_intensity = getRandomWithinPercent(intensity, 0.10);
seq_ed_wnd.setNormalizationIntensity(random_intensity);
seq_ed_wnd.synthesizeMassSpectra();
calc_options.setPolymerEntities(libXpertMassCore.Enums.ChemicalEntity.LEFT_END_MODIF |
libXpertMassCore.Enums.ChemicalEntity.FORCE_LEFT_END_MODIF);
seq_ed_wnd.setCalcOptions(calc_options);
random_intensity = getRandomWithinPercent(intensity, 0.10);
seq_ed_wnd.setNormalizationIntensity(random_intensity);
seq_ed_wnd.synthesizeMassSpectra();
// Now perform the calculation without the Phosphorylation.
calc_options.setMonomerEntities(libXpertMassCore.Enums.ChemicalEntity.NONE);
seq_ed_wnd.setCalcOptions(calc_options);
random_intensity = getRandomWithinPercent(intensity, 0.10);
seq_ed_wnd.setNormalizationIntensity(random_intensity);
seq_ed_wnd.synthesizeMassSpectra();
}
if (aborted)
break;
}
if (aborted)
break;
}The output of this script is a mass spectrum that is automatically rendered in a MineXpert program's mass spectra window if the program is installed on the computer. See Figure 7.2, “Mass spectrum as generated using the script displayed in MineXpert” .